During my PhD I wanted to build up some more of my coding skills and learn SQL. I reached out to one of my colleagues, Dr Katie Marwick, who was planning to work with the SAIL Data Bank in Wales on an epidemiological project on post-partum psychosis (PPP), to offer help with the code.
This has been a really rewarding experience. I've built up my skills in SQL, worked inside a Trusted Research Environment (a secure remote desktop where the data is held) and problem solved how to approach and clean many tricky nuances of real world data.
Below is a summary outlining the data cleaning process/pipeline and some of the interesting problems we encountered.
Where the data comes from
 {:style="display:block; margin-left:auto; margin-right:auto"}
{:style="display:block; margin-left:auto; margin-right:auto"}
This project used data from the SAIL Data Bank. Which stands for Secure Anonymised Information Linkage. This is a safe haven for anonymised person-based data of the population of Wales for research to improve health, well-being and services.
The data is structured as a database, made up of many tables that can be queried in SQL (DB2). A list of all the tables available is available on the HDR UK Gateway website. We requested access to the tables below (amongst some others, such as GP data) to extract specific variables.
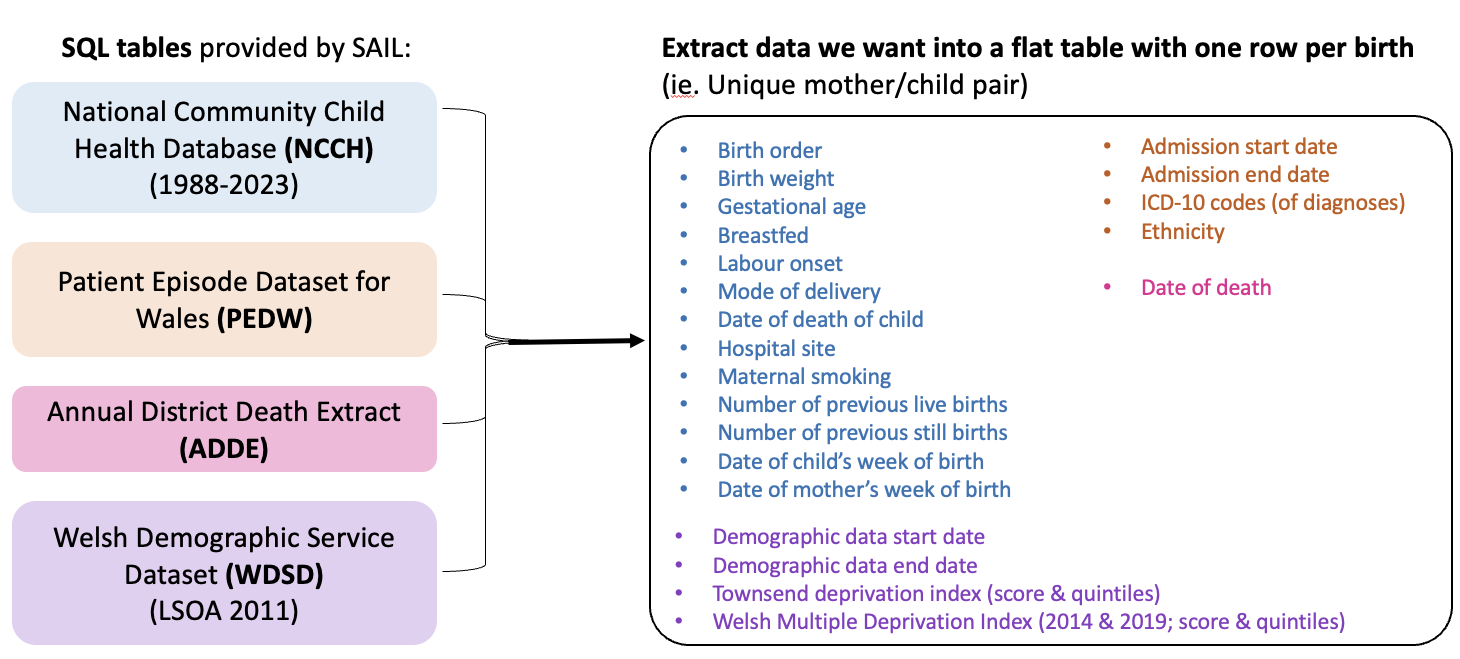 |
|---|
| Tables requested from the SAIL Data Bank, and examples of some of the variables extracted. |
Extracting the data we need
We want to extract the variables we want from those SQL tables into one table where we have one row per birth (ie. unique mother/child pair). However, it's not quite that simple as working with data on mothers and children is very nested.
The image below demonstrates this for example dummy data on one mother. Highlighting that each mother can have:
- multiple deliveries throughout her life
- multiple births per delivery (eg. twins, triplets, etc.)
- multiple hospital admissions over time (with multiple diagnoses (ICD-10 codes) per visit)
- multiple demographic data entries for different time periods throughout her life
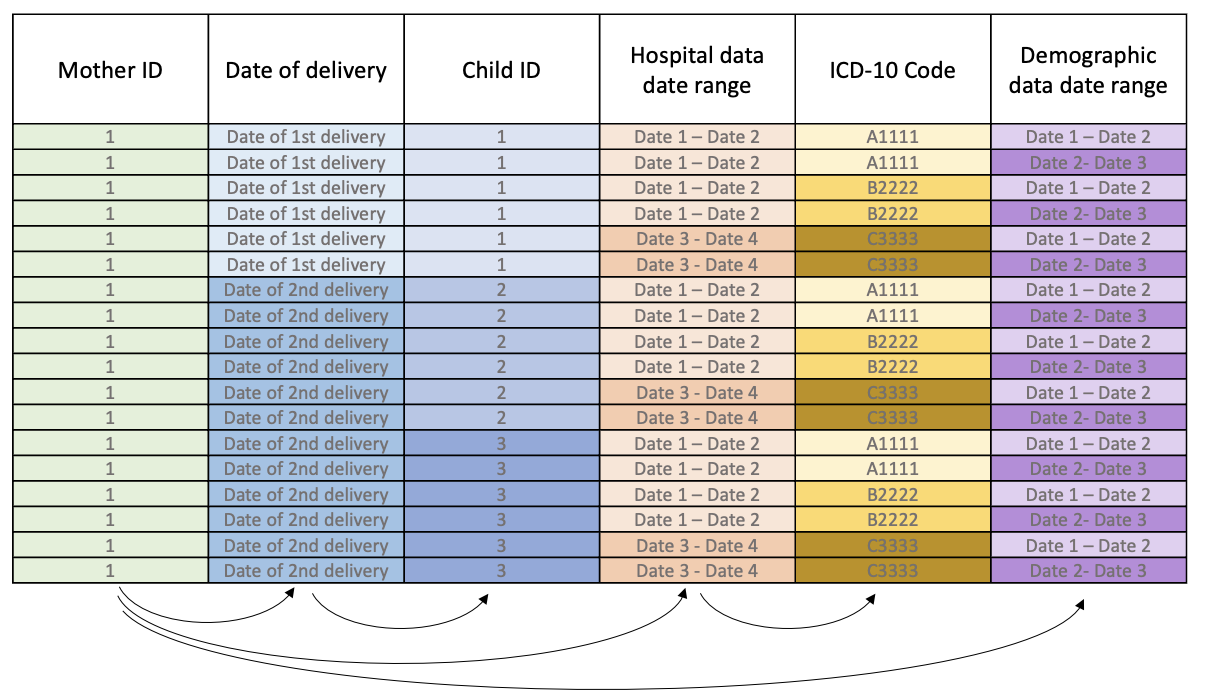 |
|---|
| Example dummy data of one mother highlighting the nesting structure of some variables. |
Some ways to reduce this nesting include:
- creating new columns for case/not case for:
- whether a mother meets the criteria for a post-partum psychosis diagnosis
- health co-morbidities we are interested in (eg. autoimmune conditions)
- keeping demographic data for the time period we are most interested in
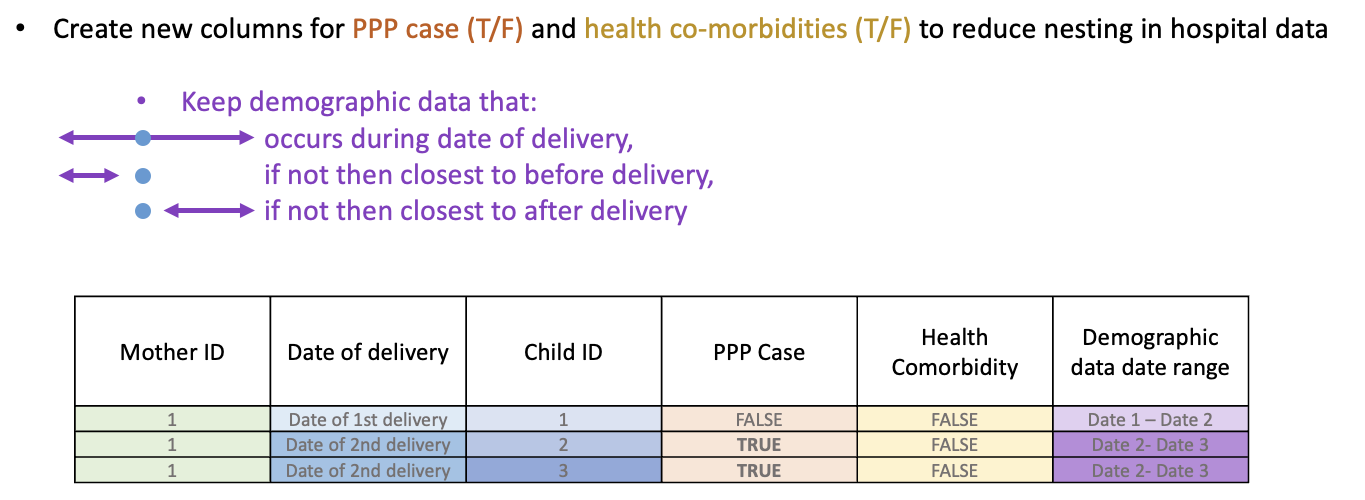 |
|---|
| Some ways to reduce most of the nesting in our table. |
However, the beady eyed amongst you may have noticed that in the above example a mother has met the criteria for a post-partum psychosis diagnosis in her second delivery but not her first. This is because criteria for post-partum psychosis is dependent on recieving a diagnosis (defined by ICD-10 codes) within a specific time window after giving birth. This adds another layer of complexity as we need to think carefully about how to handle multiple deliveries per mother. In most other papers looking at post-partum psychosis they restrict to only look at first deliveries, however doing this would reduce the number of cases and potentially overlook risk factors and co-morbidities associated with post-partum psychosis. One way to approach this is to use mixed effects model to cluster deliveries and births within mothers.
Additionally, demographic data is selected based on the most relevant entry related to time of delivery. But as already discussed a mother can have multiple demographic data entries throughout her lifetime over different time periods. This demographic data includes deprivation scores for geographical regions (Welsh Multiple Deprivation Index and Townsend Score). In the future it may also be interesting to look at other time periods per mother to capture changes as people move between different geographical regions.
Schematic of the data cleaning process
Below is an overview of the data cleaning pipeline, illustrating what the code is doing at each stage in each script.
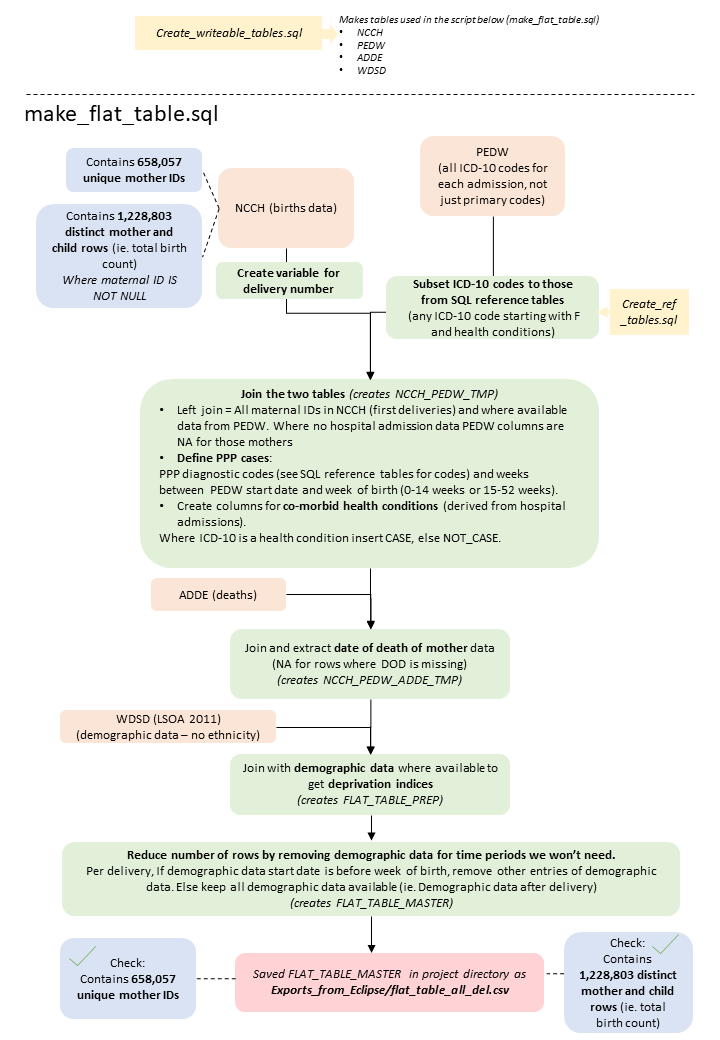
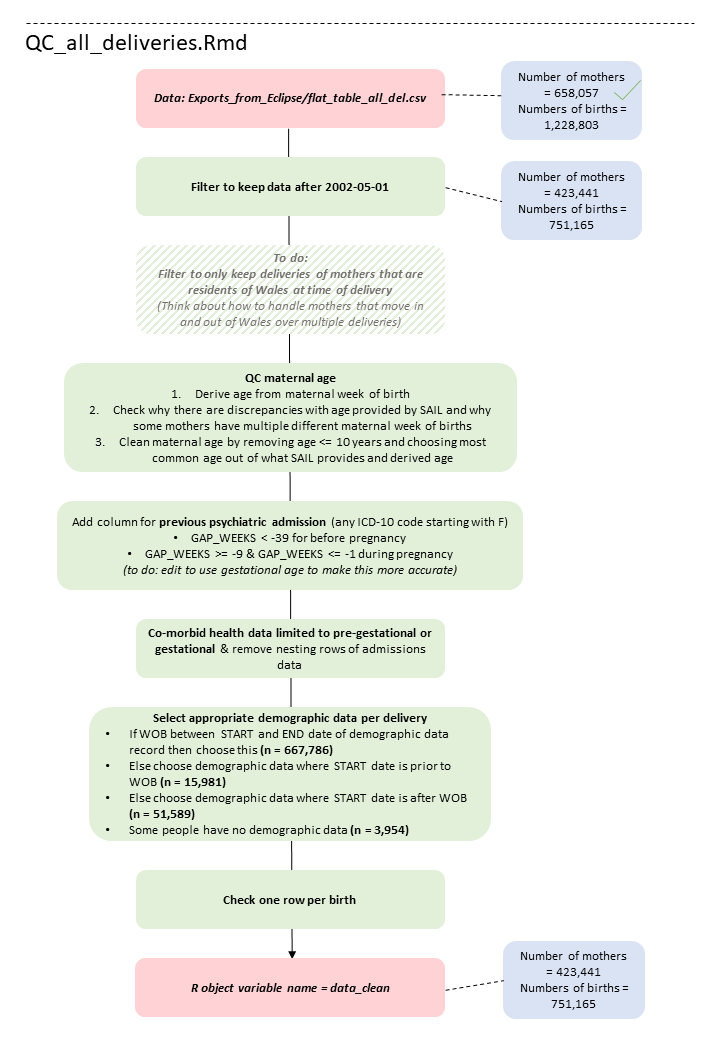
Code availability
The scripts illustrated in the figure above are available on my GitHub: https://github.com/AmeliaES/SAIL
Reflections on cleaning real world data
Hopefully I have demonstrated the complexities of working with real world health data. Some of the other issues we encountered I've not mentioned here but are detailed in the scripts linked above. These include checking variables such as maternal age (which we could check by comparing the age variable provided by SAIL against deriving an age from the mothers week of birth), then finding that some mothers had likely incorrect ages, or multiple different dates of birth.
There were also variables for previous live or still births, and to check these we could compare them with the number of previous births a mother had had. However, our dataset does not include births that occured outside of Wales (and it's possible some mothers had births in and out of Wales over a lifetime).
Additionally, when there are multiple births per delivery (eg. twins) data related to the mother or delivery was sometimes only inputted for the first child in that delivery but not subsequent. Therefore careful checking of data when there were multiple births was needed.
Overall it's been a really enjoyable challenge to work on real world, messy, health data and develop my SQL skills.
If you would like to know more about this project or are interested in collaborating please get in touch with either myself (contact details in side panel) or Dr Katie Marwick.
Acknowledgements
- Dr Katie Marwick (Project Lead).
- Collaborators: Dr Caroline Jackson and colleagues at The Usher Institute (Edinburgh, UK).
- Finally, although the above text about data cleaning is quite dry and direct, this is data on real people and sensitive subject matters. Therefore we are very grateful for the people who's anonymised data we have used from the SAIL Data Bank for this project. More information on the data included in SAIL Data Bank is available on their website.